! pip install numpy keras jax matplotlib scikit- learn
Requirement already satisfied: numpy in /usr/local/lib/python3.10/dist-packages (2.2.2)
Requirement already satisfied: keras in /usr/local/lib/python3.10/dist-packages (3.8.0)
Requirement already satisfied: jax in /usr/local/lib/python3.10/dist-packages (0.5.0)
Requirement already satisfied: matplotlib in /usr/local/lib/python3.10/dist-packages (3.10.0)
Requirement already satisfied: scikit-learn in /usr/local/lib/python3.10/dist-packages (1.6.1)
Requirement already satisfied: packaging in /usr/local/lib/python3.10/dist-packages (from keras) (24.2)
Requirement already satisfied: ml-dtypes in /usr/local/lib/python3.10/dist-packages (from keras) (0.5.1)
Requirement already satisfied: namex in /usr/local/lib/python3.10/dist-packages (from keras) (0.0.8)
Requirement already satisfied: rich in /usr/local/lib/python3.10/dist-packages (from keras) (13.9.4)
Requirement already satisfied: optree in /usr/local/lib/python3.10/dist-packages (from keras) (0.14.0)
Requirement already satisfied: h5py in /usr/local/lib/python3.10/dist-packages (from keras) (3.12.1)
Requirement already satisfied: absl-py in /usr/local/lib/python3.10/dist-packages (from keras) (2.1.0)
Requirement already satisfied: opt_einsum in /usr/local/lib/python3.10/dist-packages (from jax) (3.4.0)
Requirement already satisfied: jaxlib<=0.5.0,>=0.5.0 in /usr/local/lib/python3.10/dist-packages (from jax) (0.5.0)
Requirement already satisfied: scipy>=1.11.1 in /usr/local/lib/python3.10/dist-packages (from jax) (1.15.1)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib) (1.3.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.10/dist-packages (from matplotlib) (4.55.8)
Requirement already satisfied: kiwisolver>=1.3.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib) (1.4.8)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/lib/python3/dist-packages (from matplotlib) (2.4.7)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.10/dist-packages (from matplotlib) (0.12.1)
Requirement already satisfied: pillow>=8 in /usr/local/lib/python3.10/dist-packages (from matplotlib) (11.1.0)
Requirement already satisfied: python-dateutil>=2.7 in /usr/local/lib/python3.10/dist-packages (from matplotlib) (2.9.0.post0)
Requirement already satisfied: threadpoolctl>=3.1.0 in /usr/local/lib/python3.10/dist-packages (from scikit-learn) (3.5.0)
Requirement already satisfied: joblib>=1.2.0 in /usr/local/lib/python3.10/dist-packages (from scikit-learn) (1.4.2)
Requirement already satisfied: six>=1.5 in /usr/lib/python3/dist-packages (from python-dateutil>=2.7->matplotlib) (1.16.0)
Requirement already satisfied: typing-extensions>=4.5.0 in /usr/local/lib/python3.10/dist-packages (from optree->keras) (4.12.2)
Requirement already satisfied: pygments<3.0.0,>=2.13.0 in /usr/local/lib/python3.10/dist-packages (from rich->keras) (2.19.1)
Requirement already satisfied: markdown-it-py>=2.2.0 in /usr/local/lib/python3.10/dist-packages (from rich->keras) (3.0.0)
Requirement already satisfied: mdurl~=0.1 in /usr/local/lib/python3.10/dist-packages (from markdown-it-py>=2.2.0->rich->keras) (0.1.2)
WARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager. It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv
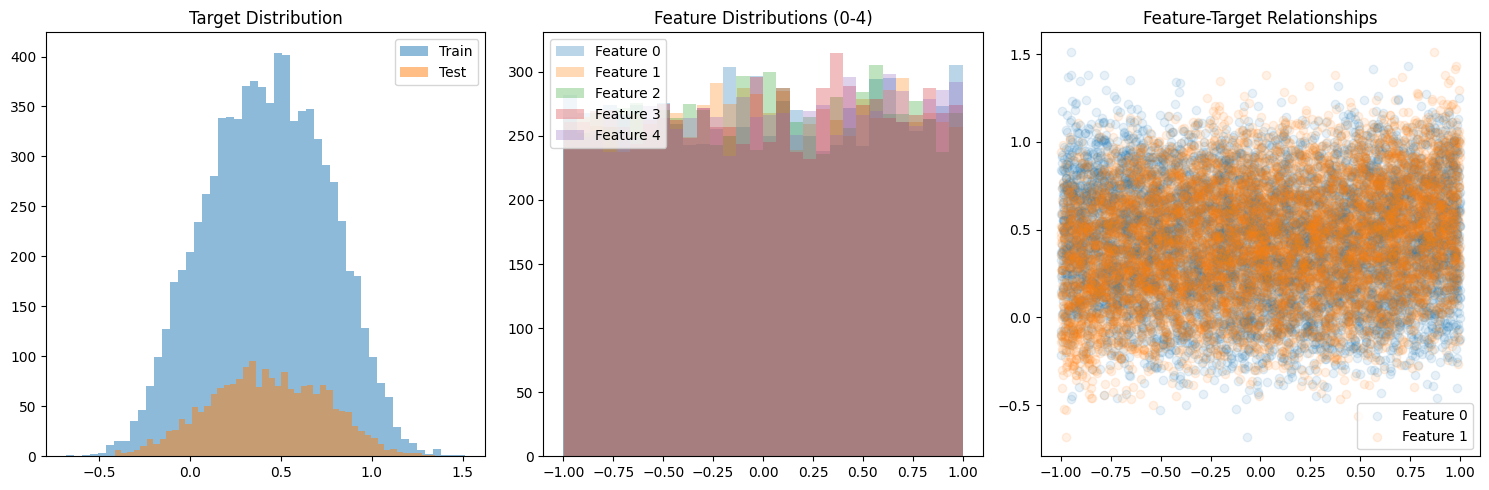
Part 0: Data Generation
We’ll use the following Feynman equation for our synthetic data: I₁₃: θ = 2π√(l/g) (Period of a pendulum)
import os"KERAS_BACKEND" ] = "jax" # Environment variable need to be set prior to importing keras, default is tensorflow import numpy as npfrom sklearn.model_selection import train_test_splitimport matplotlib.pyplot as pltdef generate_complex_data(n_samples, noise_level= 0.1 ):"""Generate synthetic data with 15 features and non-linear relationships. The target function combines: - Quadratic terms - Feature interactions - Trigonometric functions - Exponential terms """ # Generate random features = np.random.uniform(- 1 , 1 , (n_samples, 15 ))# Create complex target function = (# Polynomial terms 0.1 * X[:, 0 ]** 2 + 0.2 * X[:, 1 ]** 3 - 0.1 * X[:, 2 ] * X[:, 3 ] + # Trigonometric terms 0.3 * np.sin(2 * X[:, 4 ]) + 0.2 * np.cos(3 * X[:, 5 ]) + 0.1 * np.sin(X[:, 6 ] * X[:, 7 ]) + # Exponential terms 0.2 * np.exp(- X[:, 8 ]** 2 ) + 0.3 * np.exp(- X[:, 9 ]** 2 ) + # Linear terms with interactions 0.1 * X[:, 10 ] * X[:, 11 ] + 0.2 * X[:, 12 ] * X[:, 13 ] + # Extra feature for noise 0.1 * X[:, 14 ]# Add noise += noise_level * np.random.normal(0 , 1 , n_samples)# Reshape y and normalize = y.reshape(- 1 , 1 )return train_test_split(X, y, test_size= 0.2 , random_state= 42 )# Generate data = 10000 # More samples for higher dimensional data = generate_complex_data(n_samples)# Visualize distributions = (15 , 5 ))131 )= 50 , alpha= 0.5 , label= 'Train' )= 50 , alpha= 0.5 , label= 'Test' )'Target Distribution' )132 )for i in range (5 ): # Plot first 5 features = 30 , alpha= 0.3 , label= f'Feature { i} ' )'Feature Distributions (0-4)' )133 )0 ], y_train, alpha= 0.1 , label= 'Feature 0' )1 ], y_train, alpha= 0.1 , label= 'Feature 1' )'Feature-Target Relationships' )print ("Data shapes:" )print (f"X_train: { X_train. shape} " )print (f"X_test: { X_test. shape} " )print (f"y_train: { y_train. shape} " )print (f"y_test: { y_test. shape} " )
Data shapes:
X_train: (8000, 15)
X_test: (2000, 15)
y_train: (8000, 1)
y_test: (2000, 1)
Part 1: Basic MLP using Sequential API
Your task is to: 1. Create a baseline linear regression model using keras.Sequential with a single Dense layer 2. Create an MLP with two hidden layers (64 and 32 units) using ReLU activation 3. Compare their performance using Mean Squared Error
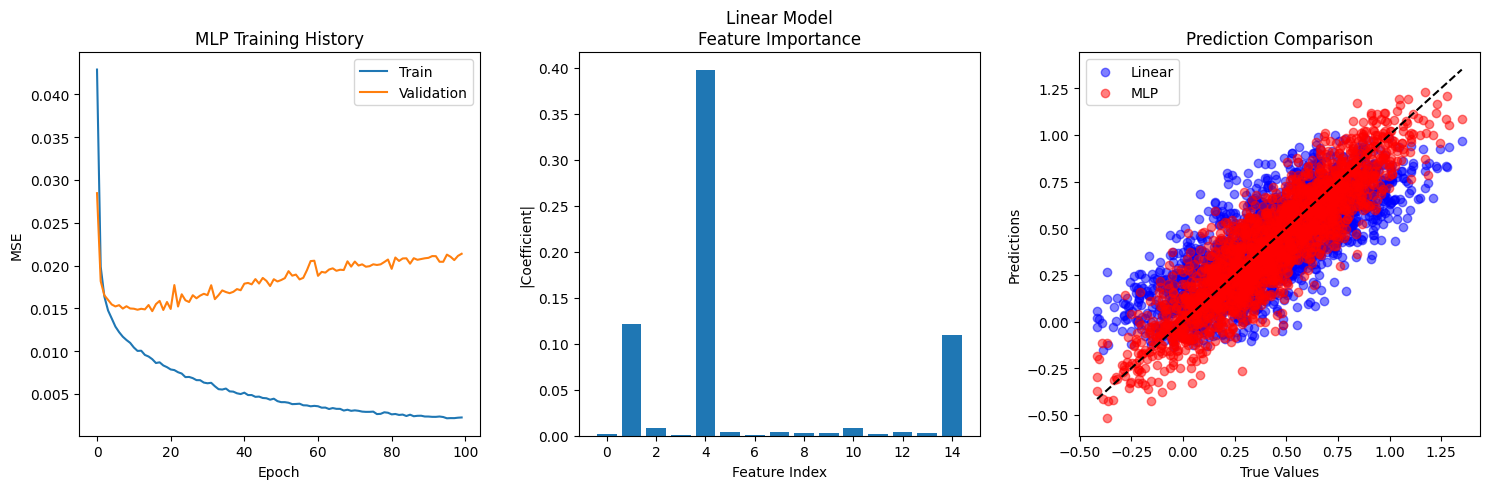
from sklearn.linear_model import LinearRegressionfrom sklearn.metrics import mean_squared_error, r2_scoreimport kerasfrom keras import layersimport keras.ops as ops# Sklearn Linear Regression baseline = LinearRegression()# Make predictions with linear model = linear_model.predict(X_train)= linear_model.predict(X_test)# Compute metrics for linear model = mean_squared_error(y_train, y_pred_linear_train)= mean_squared_error(y_test, y_pred_linear_test)= r2_score(y_train, y_pred_linear_train)= r2_score(y_test, y_pred_linear_test)# MLP with larger architecture for complex data = keras.Sequential([128 , activation= 'relu' ),64 , activation= 'relu' ),32 , activation= 'relu' ),1 )compile (optimizer= 'adam' , loss= 'mse' , metrics= ['mae' ])= mlp_model.fit(= (X_test, y_test),= 100 ,= 32 ,= 0 # Make predictions with MLP = mlp_model.predict(X_train)= mlp_model.predict(X_test)# Compute metrics for MLP = mean_squared_error(y_train, y_pred_mlp_train)= mean_squared_error(y_test, y_pred_mlp_test)= r2_score(y_train, y_pred_mlp_train)= r2_score(y_test, y_pred_mlp_test)# Print comparison print ("Model Performance Comparison:" )print (" \n Linear Regression:" )print (f"Train MSE: { linear_mse_train:.6f} " )print (f"Test MSE: { linear_mse_test:.6f} " )print (f"Train R²: { linear_r2_train:.6f} " )print (f"Test R²: { linear_r2_test:.6f} " )print (" \n MLP:" )print (f"Train MSE: { mlp_mse_train:.6f} " )print (f"Test MSE: { mlp_mse_test:.6f} " )print (f"Train R²: { mlp_r2_train:.6f} " )print (f"Test R²: { mlp_r2_test:.6f} " )# Print relative improvement = (linear_mse_test - mlp_mse_test) / linear_mse_test * 100 = (mlp_r2_test - linear_r2_test) / abs (linear_r2_test) * 100 print (f" \n Relative Improvements:" )print (f"MSE Improvement: { mse_improvement:.1f} %" )print (f"R² Improvement: { r2_improvement:.1f} %" )# MLP with larger architecture for complex data = keras.Sequential([128 , activation= 'relu' ),64 , activation= 'relu' ),32 , activation= 'relu' ),1 )compile (optimizer= 'adam' , loss= 'mse' , metrics= ['mae' ])= mlp_model.fit(= (X_test, y_test),= 100 ,= 32 ,= 0 # Compare results = (15 , 5 ))131 )'loss' ], label= 'Train' )'val_loss' ], label= 'Validation' )'MLP Training History' )'Epoch' )'MSE' )# Feature importance for linear model 132 )= np.abs (linear_model.coef_[0 ])15 ), feature_importance)'Linear Model \n Feature Importance' )'Feature Index' )'|Coefficient|' )# Prediction comparison 133 )= 0.5 , label= 'Linear' , color= 'blue' )= 0.5 , label= 'MLP' , color= 'red' )min (), y_test.max ()], [y_test.min (), y_test.max ()], 'k--' )'True Values' )'Predictions' )'Prediction Comparison' )
250/250 ━━━━━━━━━━━━━━━━━━━━ 0s 278us/step
63/63 ━━━━━━━━━━━━━━━━━━━━ 0s 836us/step
Model Performance Comparison:
Linear Regression:
Train MSE: 0.047476
Test MSE: 0.044473
Train R²: 0.566224
Test R²: 0.554408
MLP:
Train MSE: 0.002248
Test MSE: 0.020943
Train R²: 0.979464
Test R²: 0.790168
Relative Improvements:
MSE Improvement: 52.9%
R² Improvement: 42.5%
Part 2: Functional API Implementation
Create a function that builds an MLP using the Functional API: - The function should accept a list of hidden layer sizes - Each hidden layer should use ReLU activation - The output layer should be linear (no activation)
def build_mlp(hidden_sizes, input_shape= (15 ,)):= keras.layers.Input(shape= input_shape)= inputsfor units in hidden_sizes:= layers.Dense(units, activation= 'relu' )(x)= layers.Dense(1 )(x)= keras.Model(inputs= inputs, outputs= outputs)compile (optimizer= 'adam' , loss= 'mse' , metrics= ['mae' ])return model# Test functional API model = build_mlp([128 , 64 , 32 ])= func_model.fit(= (X_test, y_test),= 100 ,= 32 ,= 0 # Evaluate functional model = func_model.predict(X_train)= func_model.predict(X_test)= mean_squared_error(y_train, y_pred_func_train)= mean_squared_error(y_test, y_pred_func_test)= r2_score(y_train, y_pred_func_train)= r2_score(y_test, y_pred_func_test)print (" \n Functional API Model Performance:" )print (f"Train MSE: { func_mse_train:.6f} " )print (f"Test MSE: { func_mse_test:.6f} " )print (f"Train R²: { func_r2_train:.6f} " )print (f"Test R²: { func_r2_test:.6f} " )
250/250 ━━━━━━━━━━━━━━━━━━━━ 0s 263us/step
63/63 ━━━━━━━━━━━━━━━━━━━━ 0s 902us/step
Functional API Model Performance:
Train MSE: 0.001665
Test MSE: 0.020820
Train R²: 0.984788
Test R²: 0.791393
Part 3: Subclassing Implementation
Create a subclass of keras.Model that implements the same MLP architecture: - Should accept list of hidden sizes in init - Define layers in init - Implement forward pass in call()
class SubclassedMLP(keras.Model):def __init__ (self , hidden_sizes):super ().__init__ ()self .hidden_layers = []for units in hidden_sizes:self .hidden_layers.append(layers.Dense(units, activation= 'relu' ))self .output_layer = layers.Dense(1 )def call(self , inputs):= inputsfor layer in self .hidden_layers:= layer(x)return self .output_layer(x)# Test subclassed model = SubclassedMLP([128 , 64 , 32 ])compile (optimizer= 'adam' , loss= 'mse' , metrics= ['mae' ])= subclass_model.fit(= (X_test, y_test),= 100 ,= 32 ,= 0 # Evaluate subclassed model = subclass_model.predict(X_train)= subclass_model.predict(X_test)= mean_squared_error(y_train, y_pred_subclass_train)= mean_squared_error(y_test, y_pred_subclass_test)= r2_score(y_train, y_pred_subclass_train)= r2_score(y_test, y_pred_subclass_test)print (" \n Subclassed Model Performance:" )print (f"Train MSE: { subclass_mse_train:.6f} " )print (f"Test MSE: { subclass_mse_test:.6f} " )print (f"Train R²: { subclass_r2_train:.6f} " )print (f"Test R²: { subclass_r2_test:.6f} " )
250/250 ━━━━━━━━━━━━━━━━━━━━ 0s 216us/step
63/63 ━━━━━━━━━━━━━━━━━━━━ 0s 907us/step
Subclassed Model Performance:
Train MSE: 0.002079
Test MSE: 0.020748
Train R²: 0.981006
Test R²: 0.792121
Part 4: Custom Dense Layer
Implement your own dense layer by subclassing keras.layers.Layer: - Should accept units, activation, use_bias parameters - Implement build() method to create weights - Implement call() method for forward pass
class CustomDense(keras.layers.Layer):def __init__ (self , units, activation= None , use_bias= True ):super ().__init__ ()self .units = unitsself .activation = keras.activations.get(activation)self .use_bias = use_biasdef build(self , input_shape):= input_shape[- 1 ]# Initialize weights self .w = self .add_weight(= (input_dim, self .units),= 'glorot_uniform' ,= 'kernel' ,= True if self .use_bias:self .b = self .add_weight(= (self .units,),= 'zeros' ,= 'bias' ,= True def call(self , inputs):= ops.dot(inputs, self .w)if self .use_bias:= outputs + self .bif self .activation is not None := self .activation(outputs)return outputs= keras.Sequential([128 , activation= 'relu' ),64 , activation= 'relu' ),32 , activation= 'relu' ),1 )compile (optimizer= 'adam' , loss= 'mse' , metrics= ['mae' ])= custom_model.fit(= (X_test, y_test),= 100 ,= 32 ,= 0 # Evaluate custom dense model = custom_model.predict(X_train)= custom_model.predict(X_test)= mean_squared_error(y_train, y_pred_custom_train)= mean_squared_error(y_test, y_pred_custom_test)= r2_score(y_train, y_pred_custom_train)= r2_score(y_test, y_pred_custom_test)print (" \n Custom Dense Model Performance:" )print (f"Train MSE: { custom_mse_train:.6f} " )print (f"Test MSE: { custom_mse_test:.6f} " )print (f"Train R²: { custom_r2_train:.6f} " )print (f"Test R²: { custom_r2_test:.6f} " )
250/250 ━━━━━━━━━━━━━━━━━━━━ 0s 240us/step
63/63 ━━━━━━━━━━━━━━━━━━━━ 0s 826us/step
Custom Dense Model Performance:
Train MSE: 0.002145
Test MSE: 0.020742
Train R²: 0.980399
Test R²: 0.792181
Part 5: Advanced Custom Layer
Implement a custom layer that performs the following operation: Given input x, compute:
y = W₁(ReLU(x)) + W₂(ReLU(-x)) + b
where: - W₁, W₂ are learnable weight matrices - ReLU(x) = max(0, x) - b is a learnable bias vector
Mathematical formulation:
y = W₁ max(0, x) + W₂ max(0, -x) + bThis creates a “split” activation that learns different transformations for positive and negative inputs.
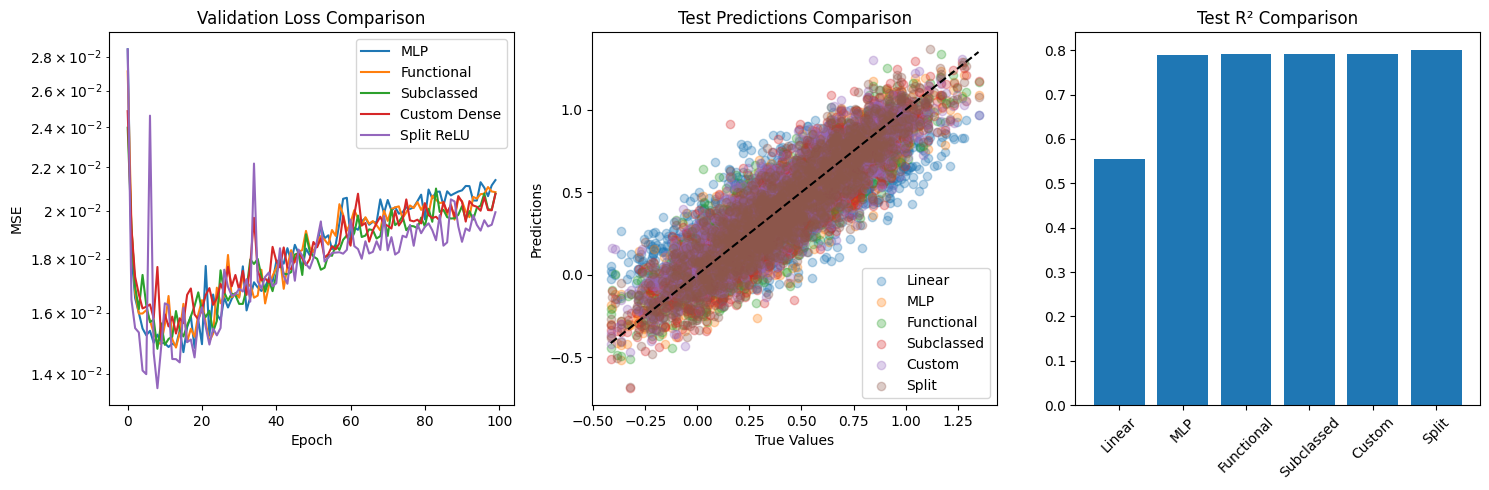
class SplitReLUDense(layers.Layer):def __init__ (self , units):super ().__init__ ()self .units = unitsdef build(self , input_shape):= input_shape[- 1 ]# Two sets of weights for positive and negative paths self .w1 = self .add_weight(= (input_dim, self .units),= 'glorot_uniform' ,= 'kernel_positive' ,= True self .w2 = self .add_weight(= (input_dim, self .units),= 'glorot_uniform' ,= 'kernel_negative' ,= True self .b = self .add_weight(= (self .units,),= 'zeros' ,= 'bias' ,= True def call(self , inputs):# Positive path = ops.dot(ops.relu(inputs), self .w1)# Negative path = ops.dot(ops.relu(- inputs), self .w2)# Combine paths return pos_path + neg_path + self .b= keras.Sequential([128 ),64 ),32 ),1 )compile (optimizer= 'adam' , loss= 'mse' , metrics= ['mae' ])= split_model.fit(= (X_test, y_test),= 100 ,= 32 ,= 0 # Evaluate split ReLU model = split_model.predict(X_train)= split_model.predict(X_test)= mean_squared_error(y_train, y_pred_split_train)= mean_squared_error(y_test, y_pred_split_test)= r2_score(y_train, y_pred_split_train)= r2_score(y_test, y_pred_split_test)print (" \n Split ReLU Model Performance:" )print (f"Train MSE: { split_mse_train:.6f} " )print (f"Test MSE: { split_mse_test:.6f} " )print (f"Train R²: { split_r2_train:.6f} " )print (f"Test R²: { split_r2_test:.6f} " )# Final comparison plot = (15 , 5 ))# Compare validation losses 131 )= {'MLP' : mlp_history,'Functional' : func_history,'Subclassed' : subclass_history,'Custom Dense' : custom_history,'Split ReLU' : split_historyfor name, history in models_histories.items():'val_loss' ], label= name)'Validation Loss Comparison' )'Epoch' )'MSE' )'log' )# Compare test predictions 132 )= {'Linear' : y_pred_linear_test,'MLP' : y_pred_mlp_test,'Functional' : y_pred_func_test,'Subclassed' : y_pred_subclass_test,'Custom' : y_pred_custom_test,'Split' : y_pred_split_testfor name, preds in models_preds.items():= 0.3 , label= name)min (), y_test.max ()], [y_test.min (), y_test.max ()], 'k--' )'Test Predictions Comparison' )'True Values' )'Predictions' )# Compare test R² scores 133 )= {'Linear' : linear_r2_test,'MLP' : mlp_r2_test,'Functional' : func_r2_test,'Subclassed' : subclass_r2_test,'Custom' : custom_r2_test,'Split' : split_r2_test= 45 )'Test R² Comparison' )
250/250 ━━━━━━━━━━━━━━━━━━━━ 0s 273us/step
63/63 ━━━━━━━━━━━━━━━━━━━━ 0s 869us/step
Split ReLU Model Performance:
Train MSE: 0.001488
Test MSE: 0.019924
Train R²: 0.986402
Test R²: 0.800373